Prepare MS data
Convert raw data files
Raw mass spectrometry data need to be converted to centroid mzML or mxXML format for MassCube data processing.
We recommend to use MSConvert for file conversion.
Visit the official website to download ProteoWizard and install MSConvert.
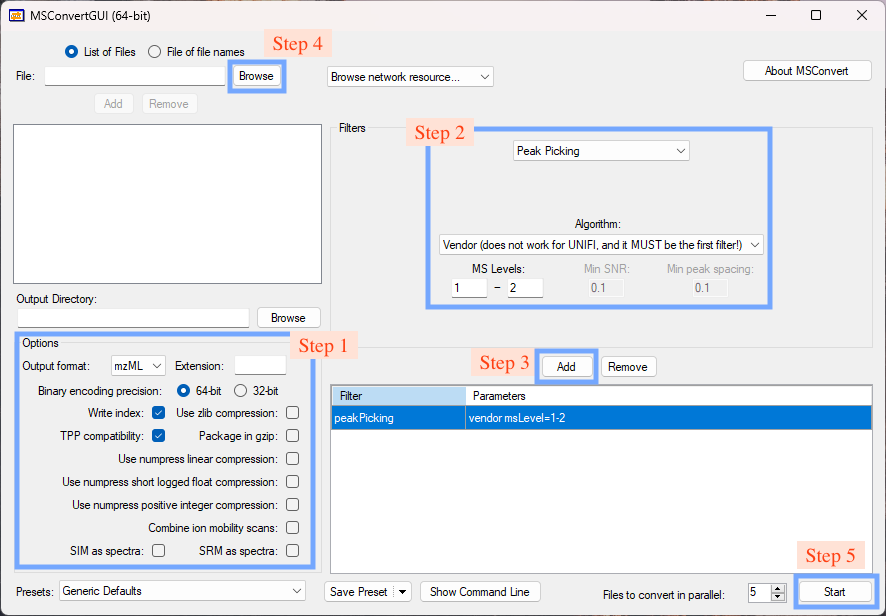
Step 1. Set options
Check the boxes as shown in Fig. 1.
Step 2. Set the peak picking filter
Step 3. Add the peak picking filter
Step 4. Browse and load files
Step 5. Start conversion
By default, the converted files will be saved in the same directory as the raw files.
Generate a sample table
In the project folder, open a terminal and run the following command:
generate-sample-tableYour project folder should include a subfolder named data that contains the raw LC-MS data files in mzML or mzXML format.
my_project
├── data
│ ├── sample1.mzML
│ ├── sample2.mzML
| └── ...
|── ...Set yes in the ‘is_blank’ column for blank samples, and yes in the ‘is_qc’ column for quality control (QC) samples. You can add more columns to the sample table to claim biological groups or other information. For example, you can add a column named ’treatment’ to claim the treatment of each sample like drug A, drug B, or control. Example:
| name | is_blank | is_qc | treatment |
|---|---|---|---|
| sample1 | no | no | drug A |
| sample2 | no | no | drug B |
| sample3 | no | no | control |
| QC1 | no | yes | NA |
| QC2 | no | yes | NA |
| blank1 | yes | no | NA |
| blank2 | yes | no | NA |